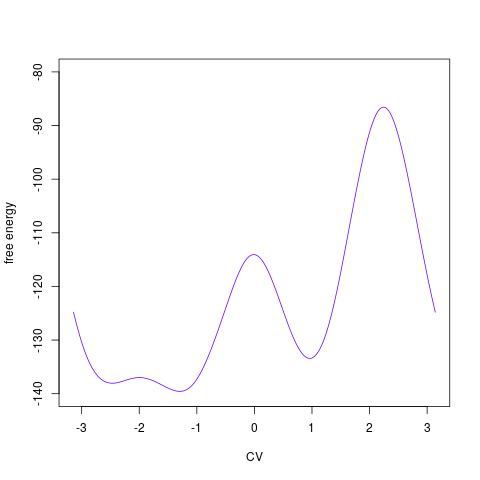
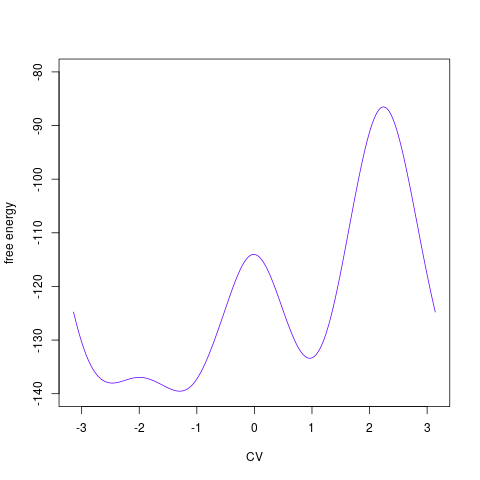
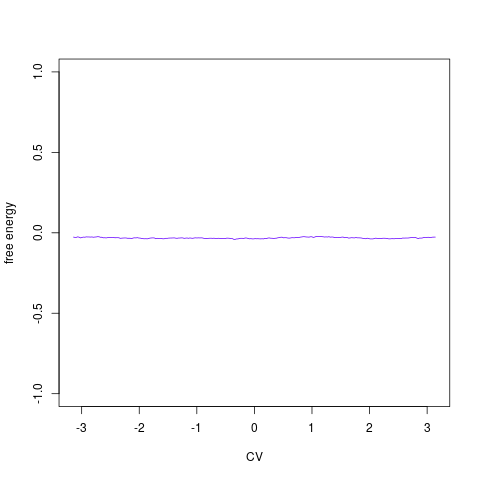
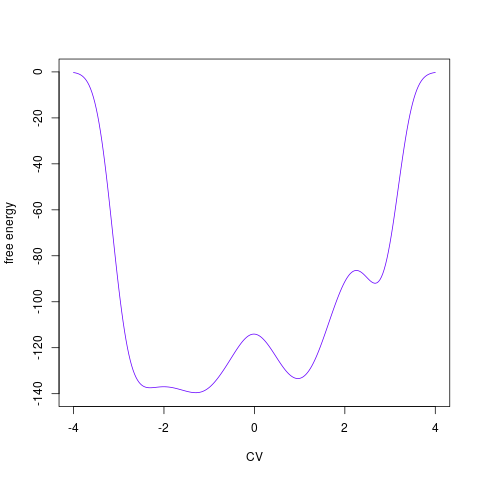
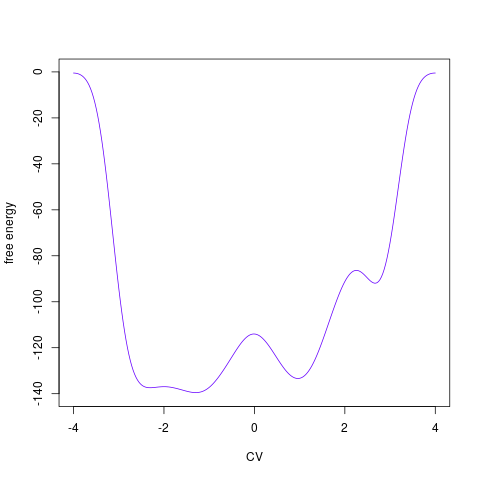
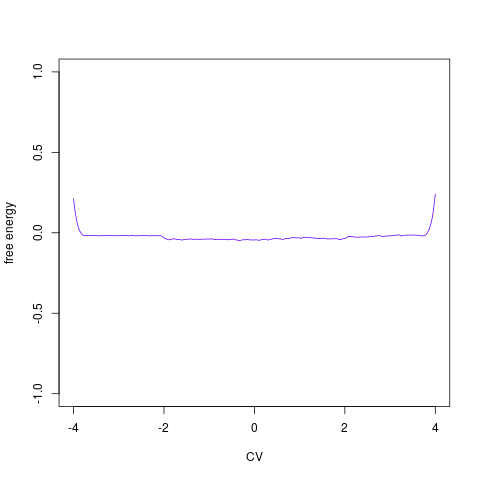
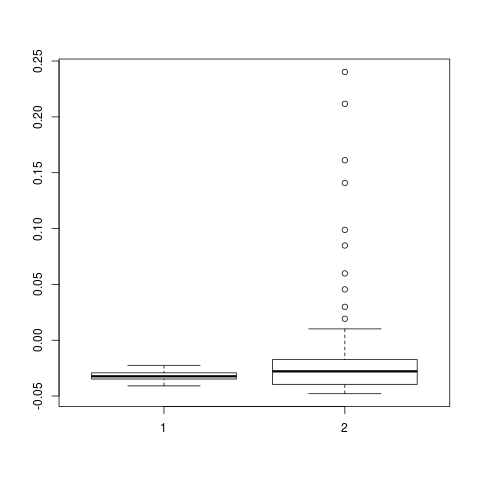
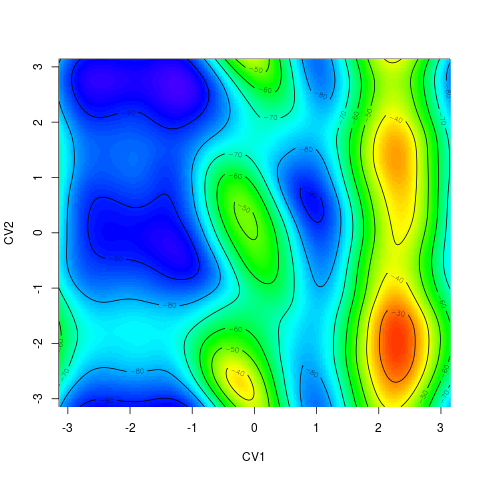
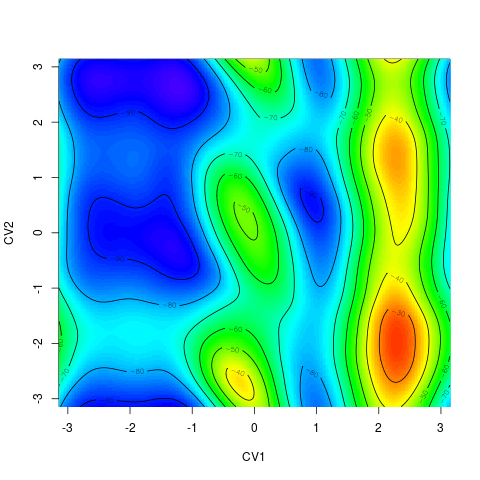
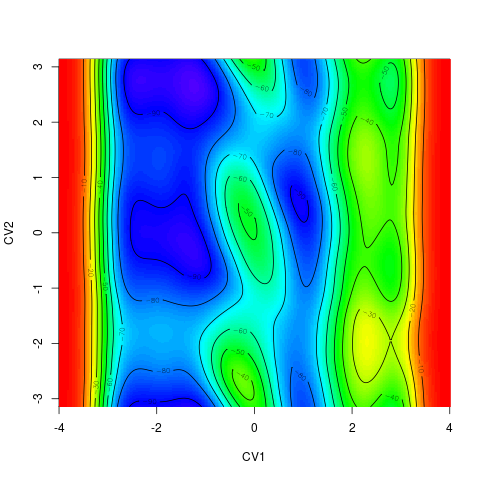
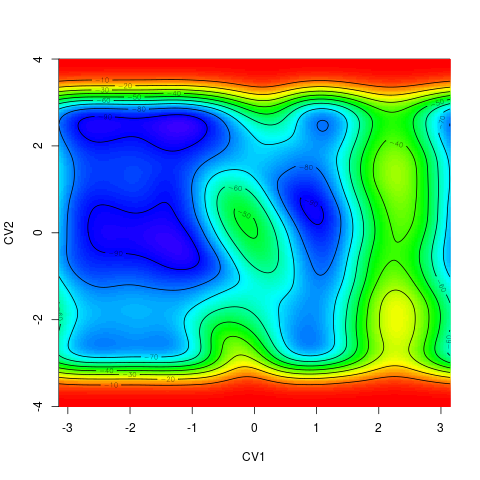
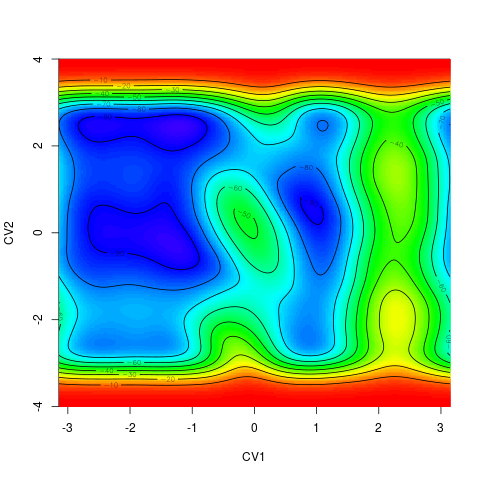
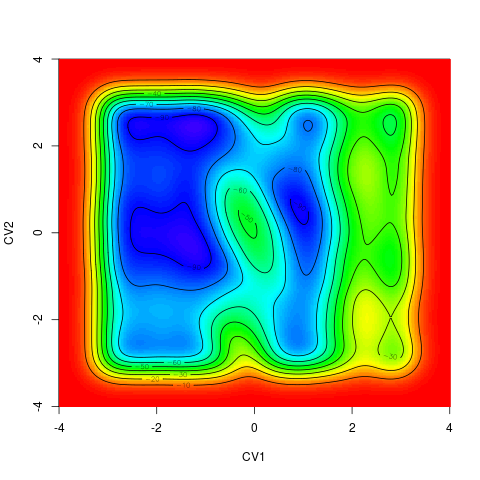
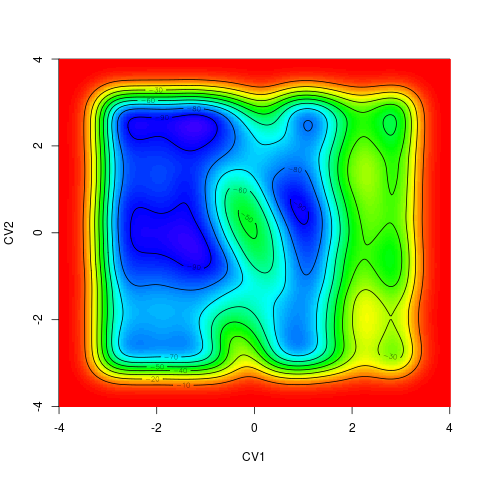
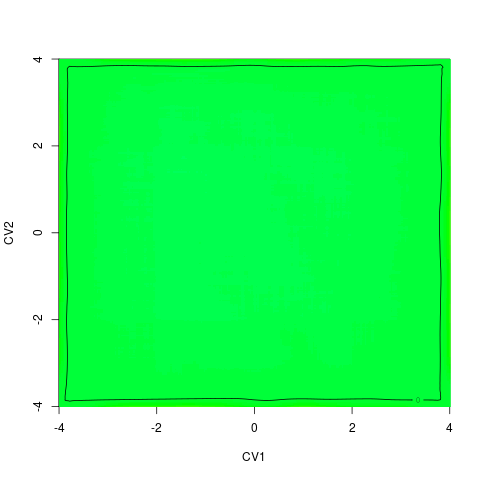
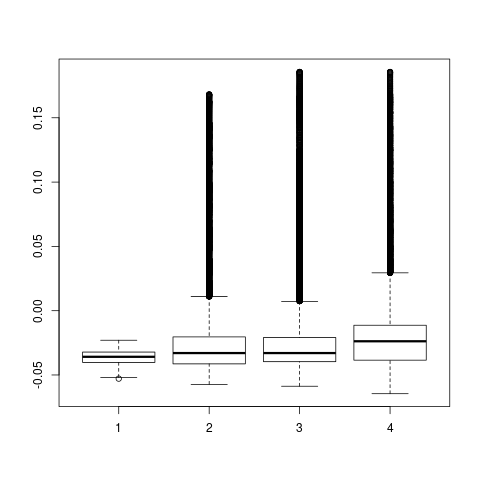
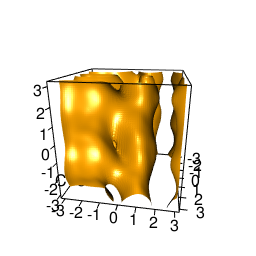
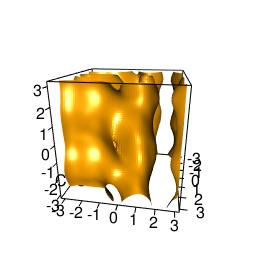
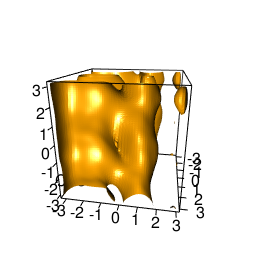
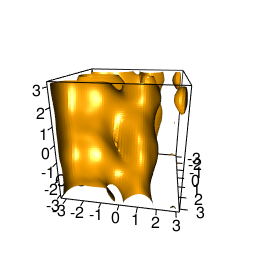
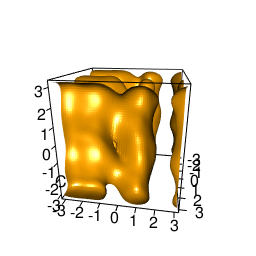
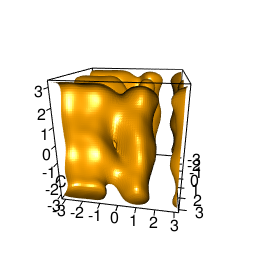
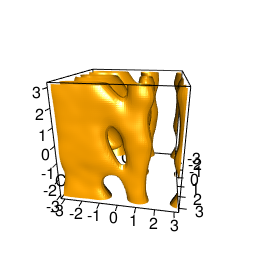
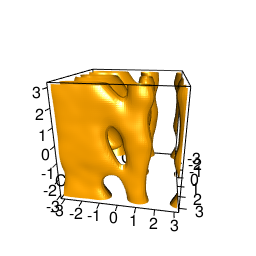
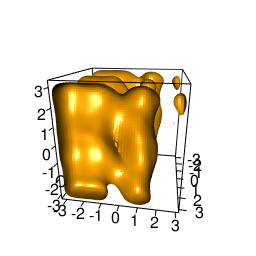
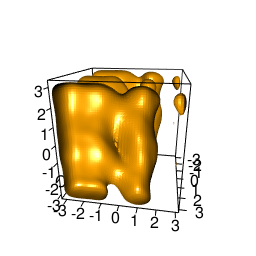
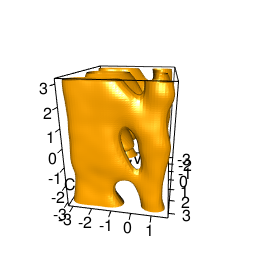
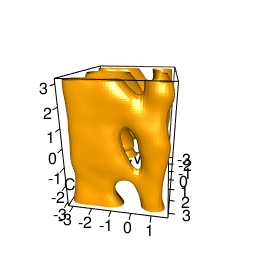
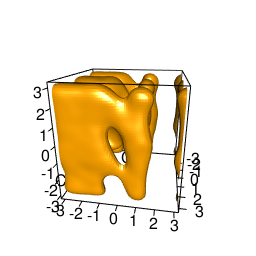
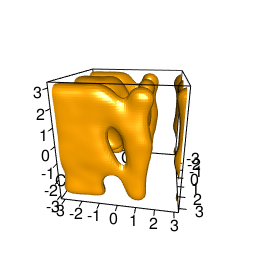
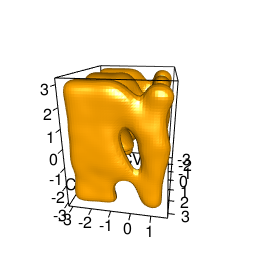
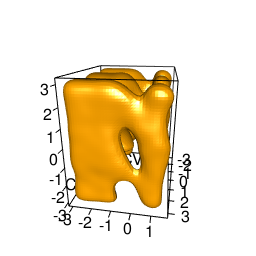
backTest of the fes2 function vs. Plumed sum_hills
Free energy surface of Alanine Dipeptide (Ace-Ala-Nme) was calculated from HILLS files by
metadynminer and Plumed
Plumed $ plumed sum_hills --hills HILLS1d --min -pi --max pi --bin 255 --outfile fes1d.txt $ plumed sum_hills --hills HILLS1dp1 --min -4 --max 4 --bin 255 --outfile fes1dp1.txt $ plumed sum_hills --hills HILLS2d --min -pi,-pi --max pi,pi --bin 255,255 --outfile fes2d.txt $ plumed sum_hills --hills HILLS2dp1 --min -4,-pi --max 4,pi --bin 255,255 --outfile fes2dp1.txt $ plumed sum_hills --hills HILLS2dp2 --min -pi,-4 --max pi,4 --bin 255,255 --outfile fes2dp2.txt $ plumed sum_hills --hills HILLS2dp12 --min -4,-4 --max 4,4 --bin 255,255 --outfile fes2dp12.txt $ plumed sum_hills --hills HILLS3d --min -pi,-pi,-pi --max pi,pi,pi --bin 63,63,63 --outfile fes3d.txt $ plumed sum_hills --hills HILLS3dp1 --min -4,-pi,-pi --max 4,pi,pi --bin 63,63,63 --outfile fes3dp1.txt $ plumed sum_hills --hills HILLS3dp2 --min -pi,-4,-pi --max pi,4,pi --bin 63,63,63 --outfile fes3dp2.txt $ plumed sum_hills --hills HILLS3dp3 --min -pi,-pi,-4 --max pi,pi,4 --bin 63,63,63 --outfile fes3dp3.txt $ plumed sum_hills --hills HILLS3dp12 --min -4,-4,-pi --max 4,4,pi --bin 63,63,63 --outfile fes3dp12.txt $ plumed sum_hills --hills HILLS3dp13 --min -4,-pi,-4 --max 4,pi,4 --bin 63,63,63 --outfile fes3dp13.txt $ plumed sum_hills --hills HILLS3dp23 --min -pi,-4,-4 --max pi,4,4 --bin 63,63,63 --outfile fes3dp23.txt $ plumed sum_hills --hills HILLS3dp123 --min -4,-4,-4 --max 4,4,4 --bin 63,63,63 --outfile fes3dp123.txtNext, metadynminer and metadynminer3d was used to obtain images of free energy surfaces, differences etc. See testit.R and testit3d.R for details. They can be run as: $ R --no-save < testit.R $ R --no-save < testit3d.R Results
|