
ContentsNews
Metadynminer and Metadynminer3D paper out in R Journal: Videos from Metadynminer Plumed Masterclass (February 2022) are available at Youtube (thanks to Plumed!!!): Video from Metadynminer webinar (31 May 2019) is available at Youtube (thanks to ELIXIR Slovenia!!!): metadynminer3d has been published.Introductionmetadynminer3d is an addendum to the package metadynminer. metadynminer is R packages for reading, analysis and visualization of metadynamics HILLS files produced by Plumed[1]. metadynminer3d visualizes 3D free energy surfaces by rgl package. It reads HILLS files from Plumed, calculates free energy surfaces by fast Bias Sum algorithm [2] and finds minima.SourceUsageInstalationTo install R see http://www.metadynamics.cz/metadynminer/index.html#par3.1.Install a stable version of metadynminer3d from CRAN by starting the R environment and typing: > install.packages("metadynminer3d")metadynminer is installed together with metadynminer3d, unless already installed. If you want to install a development version of the package from github, you need to install devtools: > install.packages("devtools") > library(devtools) > devtools::install_github("spiwokv/metadynminer3d") Next, invoke metadynminer: > library(metadynminer3d) Loading required package: metadynminer Loading required package: rglmetadynminer is loaded together with metadynminer3d, unless already loaded.
Help can be obtained by the function > help(read.hills3d) > ?read.hills Reading HILLS filesmetadynminer3d is used in similar way to metadynminer. The main difference is in loading of hills file. This is done by functionread.hills3d (see difference from read.hills in metadynminer).
A Plumed HILLS file can be loaded by typing:
> hillsf<-read.hills3d("HILLS") 3D HILLS file readThis assumes that the hills file is in the working directory. If not you can specify the path (e.g. "../mymetadynamics/HILLS"). Alternatively you can check
path by typing:
> getwd() [1] "/home/yourlogin/somedirectory"and you can change working directory by typing: > setwd("/home/yourlogin/directory")(may be useful in MS Windows). Hills file can be also loaded from a web site by using its URL: > hillsf<-read.hills3d("http://www.metadynamics.cz/metadynminer/data/HILLS3d") 3D HILLS file read metadynminer3d accepts only hills files with three collective variables (for one or two collective variables use metadynminer).
If some of collective variables is periodic (usually a torsion) it is necessary to specify
this fact at the moment of loading of hills file by parameter > hillsf<-read.hills3d("HILLS", per=c(TRUE, TRUE, TRUE)) 3D HILLS file readCollective variables are not periodic by default.
Period of a periodic collective variable is by default set to -π to +π. It can be
changed by setting parameters > hillsf<-read.hills3d("HILLS", per=c(TRUE, TRUE, TRUE), pcv1=c(0,2*pi)) 3D HILLS file readmetadynminer3d contains a preloaded data set for Alanine Dipeptide in water with Ramachandran angles φ and ψ and angle ω as collective variables (called acealanme3d).
Metadynamics simulations are very often split into multiple runs with
However, a user can load hills from restarted simulations without this option. First, wrong time values
do not affect calculations of free energy surfaces. Second, the option
The package metadynminer3d provides > acealanme3d 3D hills file HILLS2d with 30000 lines > summary(acealanme3d) 3D hills file HILLS3d with 30000 lines The CV1 ranges from -3.141557 to 3.141574 The CV2 ranges from -3.141248 to 3.141401 The CV3 ranges from -3.141561 to 3.141578Hills file objects can be concatenated by addition ( +) operation:
> acealanme3d+acealanme3d 3D hills file HILLS3d with 60000 linesmetadynminer3d provides plot function for its objects. Hills file with three collective variable will be pllotted as a 3D scatter plot of collective variable.
> plot(acealanme3d)
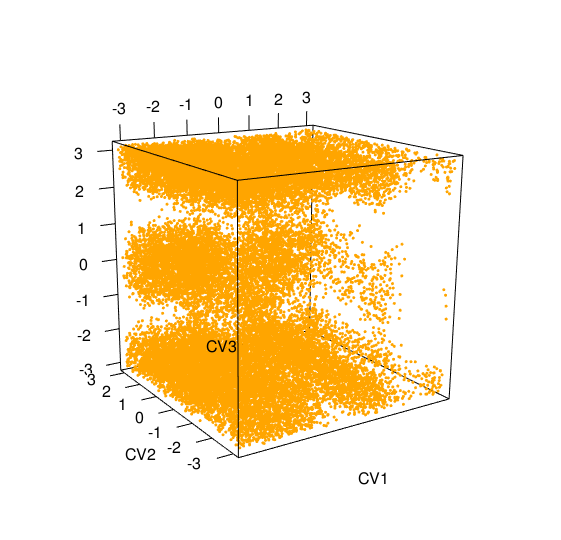
In a plot you can modify labels of axes ( In well tempered metadynamics [3] it is useful to see evolution of heights of hills. This can be plotted by typing:
> plotheights(acealanme3d)
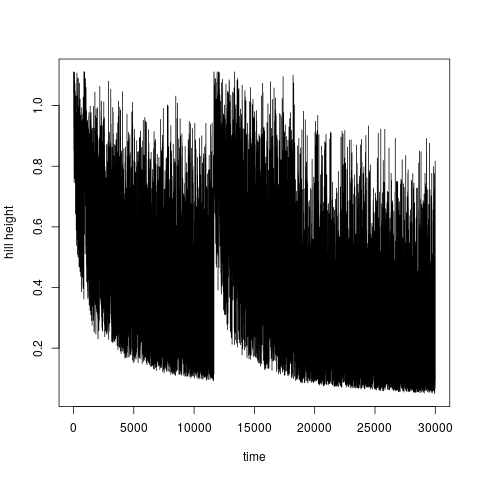
It is again possible to use Calculation of free energy surfaceFree energy surface is calculated by summing hills using functionfes:
> tfes<-fes(hillsf)Summing hills is much less expensive than a metadynamics simulation, nevertheless, it can be a slow process if number of hills exceeds 10,000. We developed a Bias Sum algorithm [2] to accelerate this calculation. Instead of evaluation exp(-||s-St||2/2σ2) for every hill it calculates a single Gaussian hill and relocates it. This function is not available for simulations with variable hill widths. As an alternative it is possible to calculate free energy surface by conventional way (function fes2). This function works with variable hill widths:
> tfes<-fes2(hillsf)Hills can be summed for whole simulation (default) or for selected hills. The range can be controlled by imin and imax. These are indexes of first and last hill to be summed.
> tfes<-fes(hillsf, imin=5000, imax=10000)By default the free energy surface is calculated for 64x64x64 grid points. This can be changed by npoints option.
Two free energy surfaces can be added and subtracted: > tfes 3D free energy surface with 64 x 64 x 64 points, maximum -0.0005099049 and minimum -77.69544 > tfes+tfes 3D free energy surface with 256 x 256 points, maximum -0.00101981 and minimum -155.3909 > tfes-tfes Warning: FES obtained by subtraction of two FESes will inherit hills only from the first FES 3D free energy surface with 64 x 64 x 64 points, maximum 0 and minimum 0It is possible to add or subtract a constant. Free energy surface can be also multiplied or divided by a constant: > tfes/4.184 Warning: division of FES will divide the FES but not hill heights 3D free energy surface with 64 x 64 x 64 points, maximum -0.0001218702 and minimum -18.56966It is possible to calculate minimum and maximum of free energy surface by functions min and
max, respectively. This can be used, for example, to convert a free energy surface so that
the global minimum is equal to zero:
> tfes-min(tfes) 3D free energy surface with 64 x 64 x 64 points, maximum 77.69493 and minimum 0The function summary function gives the same output as print for free energy
surface object:
tfes 3D free energy surface with 64 x 64 x 64 points, maximum -0.0005099049 and minimum -77.69544 summary(tfes) 3D free energy surface with 64 x 64 x 64 points, maximum -0.0005099049 and minimum -77.69544 Plotting of free energy surfaceFree energy surface can be plotted by functionplot:
> tfes<-fes(acealanme3d) > plot(tfes, level=-40) 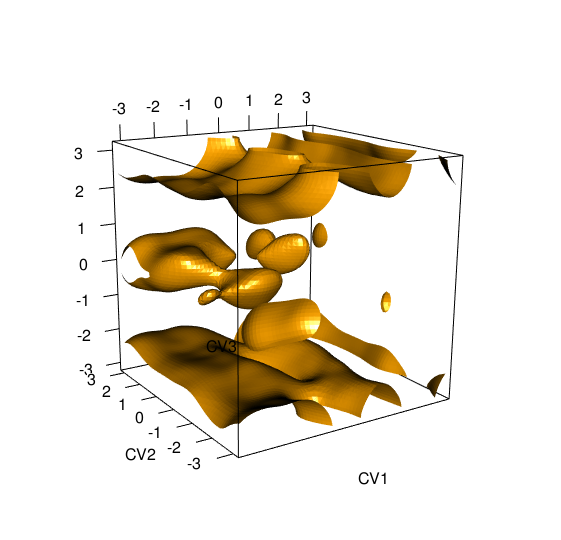
It is plotted as an isosurface at energy given by value Identification of minimaFree energy minima corresponds to stable or metastable states of the studied system. You can use functionfesminima to automatically locate free energy minima
on the free energy surface:
> tfes<-fes(acealanme3d) > minima<-fesminima(tfes)This function divides the free energy surface into 8x8x8 equivalent bins. For each bin it finds its energy minimum. Next, it checks whether these minima are local minima of the whole surface. These minima are collected, sorted and labeled by alphabet letters. The number of bins can be modified by parameter nbins.
Functions > minima $minima letter CV1bin CV2bin CV3bin CV1 CV2 CV3 free_energy 1 A 19 59 64 -1.3463969 2.6429272 3.14159265 -77.69544 2 B 7 60 64 -2.5431941 2.7426603 3.14159265 -77.01237 3 C 19 30 1 -1.3463969 -0.2493328 -3.14159265 -75.45002 4 D 42 37 1 0.9474645 0.4487990 -3.14159265 -72.31305 5 E 43 64 1 1.0471976 3.1415927 -3.14159265 -65.68216 6 F 5 60 33 -2.7426603 2.7426603 0.04986655 -61.81688 7 G 21 60 32 -1.1469307 2.7426603 -0.04986655 -61.53924 8 H 41 43 33 0.8477314 1.0471976 0.04986655 -52.84217 9 I 7 45 33 -2.5431941 1.2466638 0.04986655 -52.56641 10 J 20 26 31 -1.2466638 -0.6482652 -0.14959965 -50.88729 11 K 44 60 32 1.1469307 2.7426603 -0.04986655 -50.27426 12 L 6 25 31 -2.6429272 -0.7479983 -0.14959965 -40.22922 13 M 42 12 33 0.9474645 -2.0445286 0.04986655 -23.45578 > summary(minima) letter CV1bin CV2bin CV3bin CV1 CV2 CV3 free_energy 1 A 19 59 64 -1.3463969 2.6429272 3.14159265 -77.69544 2 B 7 60 64 -2.5431941 2.7426603 3.14159265 -77.01237 3 C 19 30 1 -1.3463969 -0.2493328 -3.14159265 -75.45002 4 D 42 37 1 0.9474645 0.4487990 -3.14159265 -72.31305 5 E 43 64 1 1.0471976 3.1415927 -3.14159265 -65.68216 6 F 5 60 33 -2.7426603 2.7426603 0.04986655 -61.81688 7 G 21 60 32 -1.1469307 2.7426603 -0.04986655 -61.53924 8 H 41 43 33 0.8477314 1.0471976 0.04986655 -52.84217 9 I 7 45 33 -2.5431941 1.2466638 0.04986655 -52.56641 10 J 20 26 31 -1.2466638 -0.6482652 -0.14959965 -50.88729 11 K 44 60 32 1.1469307 2.7426603 -0.04986655 -50.27426 12 L 6 25 31 -2.6429272 -0.7479983 -0.14959965 -40.22922 13 M 42 12 33 0.9474645 -2.0445286 0.04986655 -23.45578 relative_pop pop 1 3.376498e+13 4.359308e+01 2 2.567614e+13 3.314979e+01 3 1.372429e+13 1.771908e+01 4 3.901889e+12 5.037628e+00 5 2.733340e+11 3.528944e-01 6 5.803151e+10 7.492297e-02 7 5.191820e+10 6.703024e-02 8 1.588441e+09 2.050795e-03 9 1.422182e+09 1.836142e-03 10 7.254113e+08 9.365597e-04 11 5.673389e+08 7.324765e-04 12 1.011072e+07 1.305368e-05 13 1.213841e+04 1.567159e-08The function summary calculate equilibrium populations of each minimum in percent.
Keep in mind that these values were calculated from free energies of minima points, not by
integrating populations of each free energy minimum basin. These values are dependent on
temperature and energy units. By default these are set to temp=300 and
eunit="kJ/mol". The parameter eunit can be set to "kJ/mol"
or "kcal/mol".
Sometimes it is useful to add some point in the space of collective variables to the list of automatically identified minima. This can be done by making a "fake" minimum and adding it to the list: > minima<-fesminima(tfes)+oneminimum(tfes, cv1=0, cv2=0, cv3=0) > minima letter CV1bin CV2bin CV3bin CV1 CV2 CV3 free_energy 1 A 19 59 64 -1.3463969 2.6429272 3.14159265 -77.6954404 2 B 7 60 64 -2.5431941 2.7426603 3.14159265 -77.0123738 3 C 19 30 1 -1.3463969 -0.2493328 -3.14159265 -75.4500204 4 D 42 37 1 0.9474645 0.4487990 -3.14159265 -72.3130486 5 E 43 64 1 1.0471976 3.1415927 -3.14159265 -65.6821638 6 F 5 60 33 -2.7426603 2.7426603 0.04986655 -61.8168813 7 G 21 60 32 -1.1469307 2.7426603 -0.04986655 -61.5392350 8 H 41 43 33 0.8477314 1.0471976 0.04986655 -52.8421689 9 I 7 45 33 -2.5431941 1.2466638 0.04986655 -52.5664080 10 J 20 26 31 -1.2466638 -0.6482652 -0.14959965 -50.8872905 11 K 44 60 32 1.1469307 2.7426603 -0.04986655 -50.2742608 12 L 6 25 31 -2.6429272 -0.7479983 -0.14959965 -40.2292175 13 M 42 12 33 0.9474645 -2.0445286 0.04986655 -23.4557814 14 N 33 33 33 0.0000000 0.0000000 0.00000000 -0.5402724This will create the same set of minima with the additional one placed in the point {0,0,0}.
The function
> plot(minima)
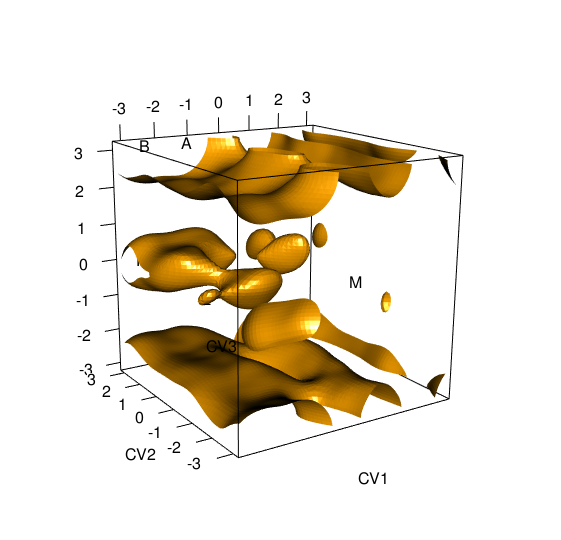
Calculation of free energy profilesConvergence can be evaluated by functionfeprof:
> tfes<-fes(acealanme3d) > minima<-fesminima(tfes) > prof<-feprof(minima)This function calculates evolution of free energy differences between each minimum and the global minimum (global at the end of simulation). Free energy of the global minimum is always zero.
Similarly to functions
It is possible to use functions > prof letter CV1bin CV2bin CV3bin CV1 CV2 CV3 free_energy 1 A 19 59 64 -1.3463969 2.6429272 3.14159265 -77.69544 2 B 7 60 64 -2.5431941 2.7426603 3.14159265 -77.01237 3 C 19 30 1 -1.3463969 -0.2493328 -3.14159265 -75.45002 4 D 42 37 1 0.9474645 0.4487990 -3.14159265 -72.31305 5 E 43 64 1 1.0471976 3.1415927 -3.14159265 -65.68216 6 F 5 60 33 -2.7426603 2.7426603 0.04986655 -61.81688 7 G 21 60 32 -1.1469307 2.7426603 -0.04986655 -61.53924 8 H 41 43 33 0.8477314 1.0471976 0.04986655 -52.84217 9 I 7 45 33 -2.5431941 1.2466638 0.04986655 -52.56641 10 J 20 26 31 -1.2466638 -0.6482652 -0.14959965 -50.88729 11 K 44 60 32 1.1469307 2.7426603 -0.04986655 -50.27426 12 L 6 25 31 -2.6429272 -0.7479983 -0.14959965 -40.22922 13 M 42 12 33 0.9474645 -2.0445286 0.04986655 -23.45578 Summary to be corrected!!!
The function > tfes<-fes(acealanme) > minima<-fesminima(tfes) > prof<-feprof(minima) > summary(prof, imind=20000, imaxd=30000) Summary to be corrected!!!
Profiles can be plotted using
> plot(prof)
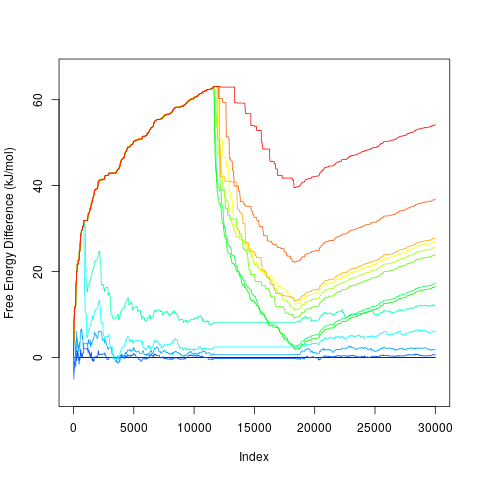
By default it plots profiles for all free energy minima. It is possible to specify minima to be
plotted by Removing one collective variableThis option is not available in metadynminer3d.Nudged Elastic BandThis option is not available in metadynminer3d.Tips and TricksPublication quality figuresFollowing script can be used to generate a publication quality figure:> tfes<-fes(acealanme3d) > plot(tfes)Change window size, zoom in and rotate if necessary, then (without closing the window) type: > rgl.snapshot(filename="plot.png") Publication of interactive FES on webYou can save free energy surface in WebGL and present it on a web site by typing:> writeWebGL(filename="index.html") Making movieIt is possible to make movie of rotation of the plot by:> movie3d(spin3d(axis=c(0,0,1)), duration=3)This will create a three-second animated GIF file of rotation around z-axis. It needs installation of convert command from ImageMagic program. Alternatively, the command:
> movie3d(spin3d(axis=c(0,0,1)), duration=3, + dir=".", convert=FALSE)These files can be concatenated by a movie making program such as mencoder. For movie of flooding look at metadynminer. Modifying aspect ratio of boxThe aspect ratio of the box can be modified by:> aspect3d(2, 1, 1)after plot command. This makes the x-axis twice longer than other axes.
References
ContactAuthor + maintenance: Vojtech Spiwok (spiwokv<you-know-what>vscht.cz)Terms of useTerms of usePrivacyPrivacy |

